Protein Aggregation Dynamics in Alzheimer's Disease
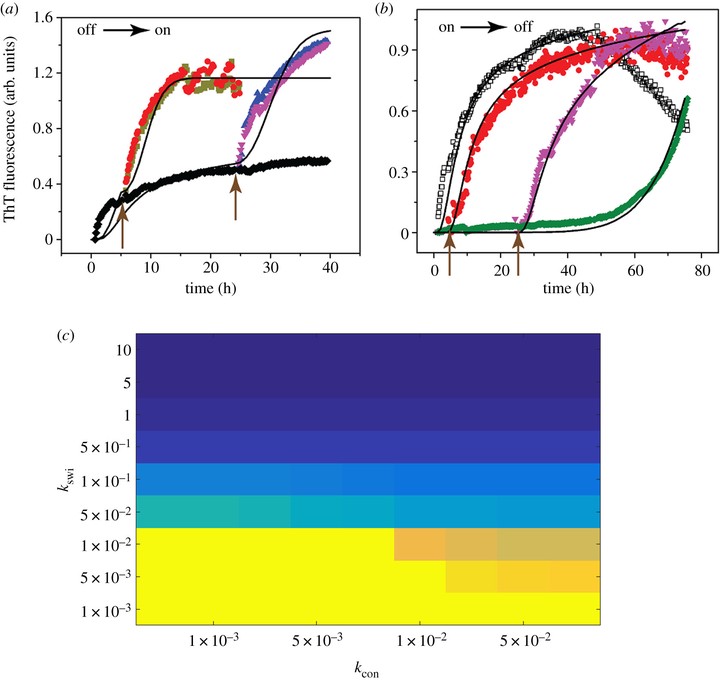 Correspondence between experimental results and EKS models on switching of pathway
Correspondence between experimental results and EKS models on switching of pathway
Alzheimer’s disease (AD) is a complex neurodegenerative disorder, clinically characterized by a gradual decline in memory and impairment of other cognitive functions like communication, movement, higher visual processing, and language inability. Polymorphism among amyloid-β (Aβ) aggregates are emerging as the main contributors to the observed phenotypic diversity in sporadic Alzheimer’s disease (sAD). Aggregation of Aβ is susceptible to several heterotypic factors, and lipids (LDs) are the most important physiological agents to interact with and modulate Aβ aggregation. Therefore, understanding the mechanism of how LDs modulate Aβ aggregation to generate polymorphic strains along competing pathways is crucial for understanding AD pathogenesis as small-sized oligomer believed to be a key cause of neural loss in AD patients. We have used the game theory model to abstract the mass-action based non-linear, coupled dynamical system of competing pathways of aggregation. Through analysis, we found higher LD concentration leads to the generation of more micelle-bound oligomers (promoting the off-pathway of aggregation), higher levels of dilution of the medium result in dissociating micelle bound oligomers enabling higher conversion to fibrils (promoting the traditional on-pathway of aggregation). These models were subsequently validated by our previously developed ensemble kinetics simulations (EKS) that use ordinary differential equations based models of the competing pathways to estimate the rate constants of converting oligomers between on- and off-pathway aggregates